Abstract
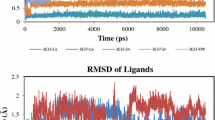
The conformational states of the peptide Cys-Val-Ile-Met (or CVIM) were computed and characterized. CVIM inhibits farnesylation of the Ras oncogene product, p21ras, at the cysteine residue of the C-terminal segment. CVIM is active in an extended conformation. A similar peptide (KTKCVFM) appears to bind the enzyme in the Type I bend conformation. In the present study, the conformations of CVIM were computed in an aqueous environment with the peptide in the zwitterionic state. Solvation free energy based on solvent accessible surface area and a distance dependent dielectric were used in the calculations. Final conformations of multiple independent Monte Carlo simulated annealing (MCSA) conformational searches were used as starting points for Metropolis Monte Carlo (MMC) runs. Conformations saved at intervals during MMC runs were analyzed. Conformers were separated by interactive clustering in dihedral angle coordinates. The four lowest energy conformers corresponding to a Type I bend, extended, AB-bend, and BA-bend were within 0.3 kcal/mol of each other, and dominant in terms of population. The Type I bend and extended conformers were supported by the binding studies. The extended conformer was the most populated. In the AB-bend conformer, `A' indicates the α-helix conformation of Val, and `B' indicates the β-strand conformation of Ile. The AB- and BA-bend conformations differed from the extended conformation in the value of Val ψ and Ile ψ, respectively, and from the Type I bend conformation in the value of Ile ψ and Val ψ, respectively. The four lowest energy conformers were characterized in terms of energy, density of low energy conformations (or entropy), structure, side chain rotamer fraction population, and interatomic distances.
Similar content being viewed by others
References
Grand, B.A. and Owen, D., Biochem. J., 279 (1991) 609.
Barbacid, M., Annu. Rev. Biochem., 56 (1987) 779.
Der, C.J. and Cox, A.D., Cancer Cells, 3 (1991) 331.
Hancock, J.F., Magee, J.E. and Marshall, C.J., Cell, 57 (1989) 1167.
Casey, P.J., Solski, P.A., Der, C.J. and Buss, J.E., Proc. Natl. Acad. Sci. USA, 86 (1989) 8323.
Gutierrez, L., Magee, A.I., Marshall, C.J. and Hancock, J.F., EMBO J., 8 (1989) 1093.
Reiss, Y., Seabra, M.C., Goldstein, J.L. and Brown, M.S., Methods, 1 (1990) 241.
Reiss, Y., Goldstein, J.L., Seabra, M.C., Casey, P.J. and Brown, M.S., Cell, 62 (1990) 81.
Leonard, D.M., J. Med. Chem., 40 (1997) 29721.
Qian, Y., Blaskovich, M.A., Saleem, M., Seong, C.M., Wathen, S.P., Hamilton, A.D. and Sebti, S.M., J. Biol. Chem., 269 (1994) 12410.
Qian, Y., Vogt, A., Sebti, S.M. and Hamilton, A.D., J. Med. Chem., 39 (1996) 217.
Leonard, D.M., Shuler, K.R., Poulter, C.J., Eaton, S.R., Sawyer, T.M., Hodges, J.C., Su, T., Scholten, J.D., Gowan, R.C., Sebolt-Leopold, J.S. and Doherty, A.M., J. Med. Chem., 39 (1996) 217.
Stradley, S.J., Rizo, J. and Gierasch, L., Biochemistry, 32 (1993) 12586.
Carlacci, L., J. Comput.-Aided Mol. Design, 12 (1998) 195.
McKelvey, D.R., Brooks, C.L. and Mokotoff, M., J. Protein Chem., 10 (1991) 265.
Zimmerman, S.S., Pottle, M.S., Némethy, G. and Scheraga, H.A., Macromolecules, 10 (1977) 1.
IUPAC-IUB Commission on Biochemical Nomenclature, Biochemistry, 9 (1970) 3471.
Karpen, M.E., Tobias, D.J. and Brooks, C.L., Biochemistry, 32 (1993) 412.
Richardson, J.S., Adv. Protein Chem., 34 (1981) 167.
Némethy, G., Pottle, M.S. and Scheraga, H.A., J. Phys. Chem., 87 (1983) 1883.
CHARMm is a trademark of Molecular Simulations Inc., San Diego, CA.
Vila, J., Williams, R.L., Vasquez, M. and Scheraga, H.A., Proteins Struct. Funct. Genet., 10 (1991) 199.
Wesson, L. and Eisenberg, D., Protein Sci., 1 (1992) 227.
Perrot, G., Cheng, B., Gibson, K.D., Vila, J., Palmer, K.A., Nayeem, A., Maigret, B. and Scheraga, H.A., J. Comput. Chem., 13 (1992) 1.
Metropolis, N., Rosenbluth, A.W., Rosenbluth, M.N., Teller, A.H. and Teller, E.J., Chem. Phys., 21 (1953) 1087.
Carlacci, L. and Englander, S.W., J. Comput. Chem., 17 (1996) 1002.
Makhatadze, G.I. and Privalov, P.L., Protein Sci., 5 (1996) 507.
Rick, S.W. and Berne, B.J., J. Am. Chem. Soc., 116 (1994) 3949.
Rick, S.W. and Berne, B.J., J. Am. Chem. Soc., 118 (1996) 672.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Carlacci, L. Conformational analysis of a farnesyltransferase peptide inhibitor, CVIM. J Comput Aided Mol Des 14, 369–382 (2000). https://doi.org/10.1023/A:1008175919794
Issue Date:
DOI: https://doi.org/10.1023/A:1008175919794