Abstract
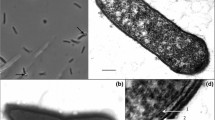
Strain T2–7, a 5-aminovalerate-fermenting bacterium previously classified as Clostridium aminovalericum, was further characterized, both physiologically and phylogenetically. Comparative sequencing analysis of the almost complete 16S rDNA revealed that strain T2–7 forms a distinct lineage within a phylogenetically coherent cluster of gram-positive bacteria currently assigned to the genus Clostridium. Strain T2–7 grew with 5-aminovalerate, 5-hydroxyvalerate, 4-hydroxybutyrate, vinylacetate, and crotonate, and required yeast extract and l-cysteine for growth. Other substrates were not utilized. The fermentation products, depending on the growth substrate, were ammonia, acetate, propionate, butyrate, and valerate. Sulphur was reduced by a mechanism not linked to energy conservation. Other acceptors were not utilized. Cells were gram-positive pointed-ended ovals, motile by means of two subpolar flagella, and possessed a gram-positive cell wall structure with an S-layer of hexagonally arranged subunits of 18.5 nm diameter. The DNA mol% G+C was 41.5. Strain T2–7 (DSM 6836) is proposed as the type strain of a new species, Clostridium viride sp. nov.
Similar content being viewed by others
References
Bader J, Günther H, Schleicher E, Simon H, Pohl S, Mannheim W (1980) Utilization of (E)-2-butenoate (crotonate) by Clostridium kluyveri and some other Clostridium species. Arch Microbiol 125:159–165
Barker HA, D'Ari L, Kahn J (1987) Enzymatic reactions in the degradation of 5-aminovalerate by Clostridium aminovalericum. J Biol Chem 262:8994–9003
Chaney AL, Marbach EP (1962) Modified reagents for determination of urea and ammonia. Clin Chem 8:130–132
Cline JD (1969) Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceangr 14:454–458
Cook GM, Janssen PH, Morgan HW (1991) Endospore formation by Thermoanaerobium brockii HTD4. System Appl Microbiol 14:240–244
Cote RJ, Gherna RL (1994) Nutrition and media. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular microbiology. American Society for Microbiology, Washington, DC, pp 155–178
De Rijk P, Neefs JM, Van de Peer Y, De Wachter R (1992) Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res 20:2075–2089
Dörner C, Schink B (1990) Clostridium homopropionicum sp. nov., a new strict anaerobe growing with 2-,3-, or 4-hydroxybutyrate. Arch Microbiol 154:342–348
Dyer JK, Costilow RM (1968) Fermentation of ornithine by Clostridium sticklandii. J Bacteriol 96:1617–1622
Eikmanns U, Buckel W (1990) Properties of 5-hydroxyvalerate CoA-transferase from Clostridium aminovalericum. Biol Chem Hoppe Seyler 371:1077–1082
Eikmanns U, Buckel W (1991a) Crystalline green 5-hydroxyvaleryl-CoA dehydratase from Clostridium aminovalericum. Eur J Biochem 197:661–668
Eikmanns U, Buckel W (1991b) A green 2,4-pentadienoyl-CoA reductase from Clostridium aminovalericum. Eur J Biochem 198:263–266
Eikmanns U, Buta C, Buckel W, Pai EF (1994) Crystallization and preliminary X-ray diffraction study of the green flavoenzyme 5-hydroxyvaleryl-CoA dehydratase/dehydrogenase from Clostridium aminovalericum. Proteins 19:269–271
Felsenstein J (1989) PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166
Gottschalk G (1986) Bacterial metabolism, 2nd edn. Springer, Berlin Heidelberg New York
Hardman JK, Stadtman TC (1960) Metabolism of ω-amino acids. II. Fermentation of δ-aminovaleric acid by Clostridium aminovalericum n. sp. J Bacteriol 79:549–552
Hippe H, Andreesen JR, Gottschalk G (1992) The genus Clostridium-nonmedical. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd edn. Springer, Berlin Heidelberg New York, pp 1800–1866
Janssen PH (1991) Isolation of Clostridium propionicum strain 19acry3 and further characteristics of the species. Arch Microbiol 155:566–571
Janssen PH, Harfoot CG (1990) Isolation of a Citrobacter species able to grow on malonate under strictly anaerobic conditions. J Gen Microbiol 136:1037–1042
Janssen PH, Morgan HW (1992) Heterotrophic sulfur reduction by Thermotoga sp. strain FjSS3.B1. FEMS Microbiol Lett 96:213–218
Johnson JL, Francis BS (1975) Taxonomy of the clostridia: ribosomal ribonucleic acid homology among the species. J Gen Microbiol 88:229–244
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, vol 3. Academic Press, New York, pp 21–132
Karn J, Brenner S, Barnett L, Cesareni G (1980) Novel bacteriophage λ cloning vector. Proc Natl Acad Sci USA 77:5172–5176
Krumböck M, Conrad R (1991) Metabolism of position-labelled glucose in anoxic methanogenic paddy soil and lake sediment. FEMS Microbiol Ecol 85:247–256
Kumar S, Tamura K, Nei M (1993) MEBA: molecular evolutionary genetics analysis, version 1.0. Pennsylvania State University, University Park, USA
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lawson PA, Llop-Perez P, Hutson RA, Hippe H, Collins MD (1993) Towards a phylogeny of the clostridia based on 16S rRNA sequences. FEMS Microbiol Lett 113:87–92
Liesack W, Bak F, Freft J-U, Stackebrandt E (1994) Holophaga foetida gen. nov., sp. nov., a new homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol 162:85–90
Lovley DR, Phillips EJP (1986) Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol 51:683–689
Magee CM, Rodeheaver G, Egerton MT, Edlich RF (1975) A more reliable Gram staining technic for diagnosis of surgical infections. Am J Surgery 130:341–346
Mead GC (1971) The amino acid-fermenting clostridia. J Gen Microbiol 67:47–56
Messner P, Hollaus F, Sleytr U (1984) Paracrystalline cell wall surface layers of different Bacillus stearothermophilus strains. Int J System Bacteriol 34:202–210
Olsen GJ, Overbeek R, Larsen N, Marsh TL, McCaughey MJ, Maciukenas MA, Kuan WM, Macke TJ, Woese CR (1992) The ribosomal data base project. Nucleic Acids Res 20:2199–2200
Pfennig N (1978) Rhodocyclus purpureus gen. nov. sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int J System Bacteriol 28:283–288
Pfennig N, Wagener S (1986) An improved method of preparing wet mounts for photomicrographs of microorganisms. J Microbiol Meth 4:303–306
Rainey FA, Ward NL, Morgan HW, Toalster R, Stackebrandt E (1993) Phylogenetic analysis of anaerobic thermophilic bacteria: aid for their reclassification. J Bacteriol 175:4772–4779
Sambrook J, Fritsch EF, Maniatis V (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Stickland LH (1935) XXXV. Studies on the metabolism of strict anaerobes (genus Clostridium). II. The reduction of proline by Cl. sporogenes. Biochem J 29:288–290
Stouthamer AH (1979) The search for correlation between theoretical and experimental growth yields. In: Quayle JR (ed) International review of biochemistry, vol 21. Microbial biochemistry. University Park Press, Baltimore, pp 1–47
Thauer RK, Jungermann K, Wenning J, Decker K (1968) Characterization of crotonate grown Clostridium kluyveri by its assimilatory metabolism. Arch Microbiol 64:125–129
Widdel F, Hansen TA (1992) The dissimilatory sulfate-and sulfurreducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd edn. Springer, Berlin Heidelberg New York, pp 583–624
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to H. A. Barker on the occasion of his 87th birthday
Rights and permissions
About this article
Cite this article
Buckel, W., Janssen, P.H., Schuhmann, A. et al. Clostridium viride sp. nov., a strictly anaerobic bacterium using 5-aminovalerate as growth substrate, previously assigned to Clostridium aminovalericum . Arch. Microbiol. 162, 387–394 (1994). https://doi.org/10.1007/BF00282102
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00282102