Abstract
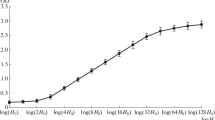
Reassociation kinetics of DNA from the macronucleus of the ciliate, Tetrahymena pyriformis GL, has been studied. The genome size determined by the kinetic complexity of DNA was found to be 2.0×108 base pairs (or 1.2×1011 daltons). About 90% of the macronuclear DNA fragments 200–300 nucleotides in length reassociate at a rate corresponding to single-copy nucleotide sequences, and 7–9% at a rate corresponding to moderate repetitive sequences; 3–4% of such DNA fragments reassociate at C0t practically equal to zero. To investigate the linear distribution of repetitive sequences, DNA fragments of high molecular weight were reassociated and reassociation products were treated with Sl-nuclease. DNA double-stranded fragments were then fractionated by size. It has been established that in the Tetrahymena genome long regions containing more than 2000 nucleotides make up about half of the DNA repetitive sequences. Another half of the DNA repetitive sequences (short DNA regions about 200–300 nucleotides long) intersperse with single-copy sequences about 1,000 nucleotides long. Thus, no more than 15% of the Tetrahymena genome is patterned on the principle of interspersing single-copy and short repetitive sequences. Most of the so called “zero time binding” or “foldback” DNA seem to be represented by inverted self-complementary (palindromic) nucleotide sequences. The conclusion has been drawn from the analysis of this fraction isolated preparatively by chromatography. About 75% of the foldback DNA is resistant to Sl-nuclease treatment. The Sl-nuclease resistance is independent of the original DNA concentration. Heat denaturation and renaturation are reversible and show both hyper and hypochromic effects. The majority of the inverted sequences are unique and about 20% are repeated tens of times. According to the equilibrium distribution in CsCl density gradients the average nucleotide content of the palindromic fraction does not differ significantly from that of total macronuclear DNA. It was shown that the largest part of this fraction of the Tetrahymena genome are not fragments of ribosomal genes.
Similar content being viewed by others
References
Allen, S.L., Gibson, J.: Genome amplification and gene expression in the ciliate macronucleus. Biochem. Genet. 6, 293–313 (1972)
Ammermann, D., Steinbrück, G., Berger, L. von, Hennig, W.: The development of the macronucleus in the ciliated protozoan Stylonichia mytilus. Chromosoma (Berl.) 33, 209–238 (1974)
Borchsenius, S.N., Belozerskaya, N.A., Merkulova, N.A., Irlina, I.S., Vorob'ev, V.I.: Possible repeated replication of a fraction of nuclear DNA in Tetrahymena during a single S-Phase. Molek. Biol. (Russ.) 11, 171–180 (1977)
Borchsenius, S.N., Belozerskaya, N.A., Merkulova, N.A., Vorob'ev, V.I.: Unique, repetitive and palindromic sequences of Tetrahymena macronuclear DNA. Molek. Biol. (Russ.) 12, 676–688 (1978)
Britten, R.J., Graham, D.E., Eden, F.C., Painchaud, D.M., Davidson, E.H.: Evolutionary divergence and length of repetitive sequences in sea urchin DNA. J. molec. Evol. 9, 1–23 (1976)
Cavalier-Smith, T.: Palindromic base sequences and replication of eukaryote chromosome ends. Nature (Lond.) 250, 467–470 (1974)
Crain, W.R., Davidson, E.H., Britten, R.J.: Contrasting patterns of DNA sequence arrangement in Apis mellifera (Honeybee) and Musca domestica (Housefly). Chromosoma (Berl.) 59, 1–12, (1976a)
Crain, W.R., Eden, F.C., Pearson, W.R., Davidson, E.H., Britten, R.J.: Absence of short period interspersion of repetitive and nonrepetitive sequences in the DNA of Drosophila melanogaster. Chromosoma (Berl.) 56, 309–326 (1976)
Davidson, E.H., Galau, G.A., Angerer, R.S., Britten, R.J.: Comparative aspects of DNA organization in metazoa. Chromosoma (Berl.) 51, 253–259 (1975)
Engberg, J., Anderson, P., Leick, V., Collins, J.: Free ribosomal DNA molecules from Tetrahymena pyriformis GL are Giant Palindromes. J. molec. Biol. 104, 455–470 (1976)
Engberg, J., Pearlman, R.E.: The amount of ribosomal RNA genes in Tetrahymena pyriformis in different physiological states. Europ. J. Biochem. 26, 393–400 (1972)
Firtel, R.A., Kindle, K.: Structural organization of the genome of the cellular slime mold: interspersion of repetitive and single-copy DNA sequences. Cell 5, 401–411 (1975)
Flamm, W.G., Bond, H.E., Burr, H.E.: Density gradient centrifugation of DNA in a fixed angle rotor. Biochim. biophys. Acta (Amst.) 129, 310–319 (1966)
Graham, D.E., Neufeld, B.R., Davidson, E.H., Britten, R.J.: Interspersion of repetitive and nonrepetitive DNA sequences in the sea urchin genome. Cell 1, 127–136 (1974)
Irlina, I.S., Merkulova, N.A.: The growing in volume and mass of Tetrahymena pyriformis for biochemical purposes and the synchronization of division. Cytology (Russ.) 17, 1208–1215 (1975)
Karrer, K.M., Gall, J.G.: The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J. molec. Biol. 104, 421–454 (1976)
Kupriyanova, N.S., Timofeeva, M.Ja., Baev, A.A.: Detection and characterization of the palindromes in a loach genome. Molek. Biol. (Russ.) 10, 412–422 (1976)
Manning, J.E., Schmidt, C.W., Davidson, N.: Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell 4, 141–155 (1975)
Perlman, S., Phillips, C., Bishop, J.O.: A study of fold-back DNA. Cell 8, 33–42 (1976)
Prensky, W.: The radioiodination of the RNA and DNA to high specific activities. In: Methods in cell biology (D.M. Prescott, ed.), vol. 13, pp. 121–152. New York-London: Academic Press 1976
Raikov, I.B.: Evolution of macronuclear organization. Ann. Rev. Genet. 10, 413–440 (1976)
Ryskov, A.P., Saunders, G.F., Farashyan, V.R., Georgiev, G.P.: Double-helical regions in nuclear precursor of m-RNA (pre-m RNA). Biochim. biophys. Acta (Amst.) 312, 152–164 (1973)
Thomas, C.A., Jr., Pyeritz, R.E., Wilson, D.A., Dancis, B.M., Lee, C.S., Bick, M.D., Huang, H.L., Zimm, B.H.: Cyclodromes and palindromes in chromosomes. Cold Spr. Harb. Symp. quant. Biol. 38, 353–370 (1974)
Vogt, V.H.: Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Europ. J. Biochem. 33, 192–200 (1973)
Wetmur, J.G., Davidson, N.: Kinetics of renaturation of DNA. J. molec. Biol. 31, 349–370 (1968)
Wilson, D.A., Thomas, C.A., Jr.: Palindromes in chromosomes. J. molec. Biol. 84, 115–138 (1974)
Yao, M.-C., Gorovsky, M.A.: Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma (Berl.) 48, 1–18 (1974)
Zimmerman, I.L., Goldberg, R.B.: DNA sequence organization in the genome of Nicotiana tabacum. Chromosoma (Berl.) 59, 227–252 (1976)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Borchsenius, S.N., Belozerskaya, N.A., Merkulova, N.A. et al. Genome structure of Tetrahymena pyriformiis . Chromosoma 69, 275–289 (1978). https://doi.org/10.1007/BF00332132
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00332132