Summary
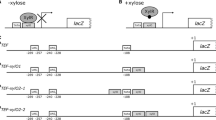
The relationship between the promoter length of the Kluyveromyces fragilis β-glucosidase gene and the level of its expression in Saccharomyces cerevisiae was studied by gene fusion between deleted promoter fragments of various lengths and the promoterless β-galactosidase gene of Escherichia coli. The removal of a region from position-425 to-232 led to a tenfold increase in the expression of the gene. The same results were obtained for the reconstructed β-glucosidase gene with the same promoter length. It is likely that the deletion of this part of the promoter removes negative regulatory elements which are functional in Saccharomyces cerevisiae. This increase in activity is the main event which may explain the high increase in gene expression (60-fold) previously observed for an upstream deletion obtained during subcloning experiments of the β-glucosidase gene. It is also shown that the expression of the gene greatly depends upon the nature of the recipient strain, the growth phase of the cell and that of the vector carrying it.
Similar content being viewed by others
References
Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein dye binding. Anal Biochem 72:248–254
Biggin MD, Gibson TJ, Hong GF (1983) Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci USA 80:3963–3965
Casadaban MJ, Martinez-Arias A, Shapira SK (1983) Beta-galactosidase gene fusion for analysing gene expression in Escherichia coli and yeast. Methods Enzymol 100:293–308
Gerbaud C, Elmerich C, Tandeau de Marsac N, Chocat P, Charpin N, Guerineau M, Aubert JP (1981) Construction of new yeast vectors and cloning of the Nif (nitrogen fixation) gene cluster of Klebsiella pneumoniae in yeast. Curr Genet 3:173–180
Gray H, Ostrander D, Hodne H, Legerski R, Robbertson D (1975) Extranuclear nucleases of Pseudomonas Bal 31. Nucleic Acids Res 2:1459–1492
Guarente L (1984) Yeast promoters: positive and negative elements. Cell 36:799–800
Herman A, Halvorson H (1963) Identification of the structural gene for beta glucosidase in Saccharomyces lactis. J Bacteriol 85:895–900
Hitzeman RA, Hagie FE, Levine HL, Goeddel DV, Ammerer G, Hall BD (1981) Expression of a human gene for interferon in yeast. Nature 293:717–722
Hu N, Messing J (1982) The making of strand specific M13 probes. Gene 17:271–277
Iborra F, Francingues MC, Guerineau M (1985) Localization of the upstream regulatory sites of yeast iso2-cytochrome c gene. Mol Gen Genet 199:117–122
Maniatis T, Fritsch F, Sambrook J (1982) Molecular cloning: a Inboratory manual. Cold Spring Harbor Laboratory Press, New York
Mellor J, Dobson MJ, Roberts NA, Kingsman AJ, Kingsman SM (1985) Factors affecting heterologous gene expression in Saccharomyces cerevisiae. Gene 33:215–226
Miller J (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, pp 352–355
Ogden JE, Stanway C, Kim S, Mellor J, Kingsman AJ, Kingsman SM (1986) Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol 6:4335–4343
Raynal A, Guerineau M (1984) Cloning and expression of the structural gene for beta-glucosidase of Kluyveromyces fragilis in Escherichia coli and Saccharomyces cerevisiae. Mol Gen Genet 195:108–115
Raynal A, Gerbaud C, Francingues MC, Guerineau M (1987) Sequence and transcription of the beta glucosidase gene of Kluyveromyces fragilis cloned in Saccharomyces cerevisiae. Curr Genet 12:175–184
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Struhl K (1987) Promoters, activator proteins and the mechanism of transcriptional initiation in yeast. Cell 49:295–297
Author information
Authors and Affiliations
Additional information
Communicated by W. Gajewski
Rights and permissions
About this article
Cite this article
Iborra, F., Raynal, A. & Guerineau, M. The promoter of the β-glucosidase gene from Kluyveromyces fragilis contains sequences that act as upstream repressing sequences in Saccharomyces cerevisiae . Molec. Gen. Genet. 213, 150–154 (1988). https://doi.org/10.1007/BF00333412
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00333412