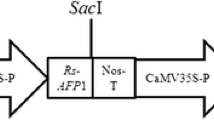
Abstract
Chitinases accumulate in higher plants upon pathogen attack are capable of hydrolyzing chitin-containing fungal cell walls and are thus implicated as part of the plant defense response to fungal pathogens. To evaluate the relative role of the predominate chitinase (class I, basic enzyme) of Arabidopsis thaliana in disease resistance, transgenic Arabidopsis plants were generated that expressed antisense RNA to the class I chitinase. Young plants or young leaves of some plants expressing antisense RNA had <10% of the chitinase levels of control plants. In the oldest leaves of these antisense plants, chitinase levels rose to 37–90% of the chitinase levels relative to vector control plants, most likely because of accumulation and storage of the enzyme in vacuoles. The rate of infection by the fungal pathogen Botrytis cinerea was measured in detached leaves containing 7–15% of the chitinase levels of control plants prior to inoculation. Antisense RNA was not effective in suppressing induced chitinase expression upon infection as chitinase levels increased in antisense leaves to 47% of levels in control leaves within 24 hours after inoculation. Leaves from antisense plants became diseased at a slightly faster rate than leaves from control plants, but differences were not significant due to high variability. Although the tendency to increased susceptibility in antisense plants suggests that chitinases may slow the growth of invading fungal pathogens, the overall contribution of chitinase to the inducible defense reponses in Arabidopsis remains unclear.
Similar content being viewed by others
References
Bartnicki-Garcia S: Cell wall chemistry, morphogenesis and taxonomy of fungi. Annu Rev Microbiol 22: 87–108 (1968).
Benhamou N, Joosten MHAJ, de Wit PJGM: Subcellular localization of chitinase and of its potential substrate in tomato root tissues infected by Fusarium oxysporum f. sp. radicis-lycopersicl. Plant Physiol 92: 1108–1120 (1990).
Benfey PN, Chua N-H: Regulated genes in transgenic plants. Science 244: 174–181 (1989).
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utlixing the principle of protein-dye binding. Anal Biochem 54: 484–489 (1976).
Broglie K, Chet I, Holliday M, Cressman R, Broglie R: Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254: 1194–1197 (1991).
Brockaert WF, Van Parijs J, Allen AK, Peumans WJ: Comparison of some molecular, enzymatic and antifungal properties of chitinases from thorn-apple, tobacco and wheat. Physiol Mol Plant Path 33: 319–331 (1988).
Collinge DB, Kragh KM, Mikkelsin JD, Kielsen KK, Rasmussen U, Vad K. Plant chitinases. Plant J 3: 31–40 (1993).
Fisher RF, Long SR: Rhizobium-plant signal exhange. Nature 357: 655–660 (1990).
Hedrick AS, Bell JN, Boller T, Lamb CJ: Chitinase cDNA cloning and mRNA induction by fungal elicitor, wounding and infection. Plant Physiol 86: 182–186 (1988).
Horsch RB, Klee HJ: Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: role of the T-DNA borders in the transfer process. Proc Natl Acad Sci USA 83: 4428–4432 (1986).
de Jong AJ, Cordewener J, Schiavo FL, Terzi M, Vandekerckhove J, van Kammen AV, de Vries SC. A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4: 425–433 (1992).
Joosten MHAJ, de Wit PJGM: Indentification of several pathogenesis-related proteins in tomato leaves inoculated with Cladosporium fulvum (syn. Fulvia fulva) as 1,3-β-glucanases and chitinases. Plant Physiol 89: 945–951 (1989).
Kilby NJ, Leyser HMO, Furner IJ: Promoter methylation and progressive transgene inactivation in Arabidopsis. Plant Mol Biol 20: 103–112 (1992).
Koch E, Slusarenko AJ: Fungal pathogens of Arabidopsis thaliana (L.) Heyhn. Bot Helv 100: 257–268 (1990).
Koncz C, Schell J: The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 (1986).
Kurosaki F, Tashiro N, Nishi A: Role of chitinase and chitin oligosaccharides in lignification response of carrot cells treated with mycelial walls. Plant Cell Physiol 29: 527–531 (1988).
Lotan T, Ori N, Fluhr R: Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1: 881–887 (1989).
Mauch F, Mauch-Mani B, Boller T: Antifungal hydrolases in pea tissue II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol 88: 936–942 (1988).
Mauch F, Staehelin LA: Functional implications of the subcellular localization of ethylene-induced chitinase and β-1,3-glucanase in bean leaves. Plant Cell 1: 447–457 (1989).
Memelink J, Linthorst JM, Schilperoort RA, Hoge JHC: Tobacco genes encoding acidic and basic isoforms of pathogenesis-related proteins display different expression patterns. Plant Mol Biol 14: 119–126 (1990).
Metraux JP, Boller T. Local and systemic induction of chitinase in cucumber plants in response to viral, bacterial and fungal infections. Physiol Mol Plant Path 28: 161–169 (1986).
Neale AD, Wahleithner JA, Lund M, Bonnett HT, Kelly A, Meeks-Wagner DR, Peacock WJ, Dennis ES: Chitinase, β-1,3-glucanase, osmotin, and extensin are expressed in tobacco explants during flower formation. Plant Cell 2: 673–684 (1990).
Neuhaus J-M, Sticher L, Meins F, Boller T: A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA 88: 10362–10366 (1991).
Neuhaus J-M, Flores S, Keefe D, Ahl-Goy P, Meins F: The function of vacuolar β-1,3-glucanase investigated by antisense transformation. Susceptibility of transgenic Nicotiana sylvestris plants to Cercospora nicotianae infection. Plant Mol Biol 19: 803–813 (1992).
Ottaviani MP, Smits T, Tencate CHH: Differential methylation and expression of the beta-glucuronidase and neomycin phosphotransferase genes in transgenic plants of potato cv. Bintje. Plant Sci 88: 73–81 (1993).
Rasmussen U, Giese H, Mikkelsen JD. Induction and purification of chitinase in Brassica napus L. ssp. oleifera infected with Phoma lingam. Planta 187: 328–334 (1992).
Roby D, Gadelle A, Toppan A: Chitin oligosaccharides as elicitors of chitinase activity in melon plants. Biochem Biophys Res Commun 143: 885–892 (1987).
Roche P, Lerouge P, Ponthus C, Promé JC: Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J Biol Chem 266: 10933–10940 (1991).
Samac DA, Hironaka CM, Yallaley PE, Shah DM: Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol 93: 907–914 (1990).
Samac DA, Shah DM: Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3: 1063–1072 (1991).
Shinshi H, Mohnen D, Meins F: Regulation of a plant pathogenesis-related enzyme: inhibition of chitinase and chitinase mRNA accumulation in cultured tobacco tissues by auxin and cytokinin. Proc Natl Acad Sci USA 84: 89–93 (1987).
Valvekens D, Van Montagu M, Van Lijsebettens M: Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 (1988).
Verberg JG, Huynh QK: Purification and characterization of an antifungal chitinase from Arabidopsis thaliana. Plant Physiol 95: 450–455 (1991).
Verberg JG, Rangwala SH, Samac DA, Luckow VA, Huynh QK: Examination of the role of tyrosine-174 in the catalytic mechanism of the Arabidopsis thaliana chitinase: comparison of variant chitinases generated by site-directed mutagenesis and expressed in insect cells using baculovirus vectors. Arch Biochem Biophys 300: 223–230 (1993).
Verhoeff K: The infection process and host-pathogen interactions. In: Coley-Smith JR, Verhoeff K, Jarvis WR (eds) The Biology of Botrytis, pp. 153–180. Academic Press, New York (1980).
Voisey CR, Slusarenko AJ: Chitinase mRNA and enzyme activity in Phaseolus vulgaris (L) increase more rapidly in response to avirulent than to virulent cells of Pseudomonas syringae pv. phaseolicola. Physiol Mol Plant Pathol 35: 403–412 (1989).
Wubben JP, Joosten MHAJ, van Kan JAL, de Wit PJGM. Subcellular localization of plant chitinases and 1,3-β-glucanases in Cladosporium fulvum (syn Fulvid fulva)-infected tomato leaves. Physiol Mol Plant Path 41: 23–32 (1992).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Samac, D.A., Shah, D.M. Effect of chitinase antisense RNA expression on disease susceptibility of Arabidopsis plants. Plant Mol Biol 25, 587–596 (1994). https://doi.org/10.1007/BF00029598
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00029598