Summary
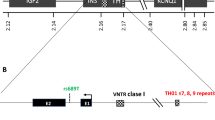
An inconsistency has come to light between the conclusion of Lucassen et al. that IDDM2 (11p15.5) must lie within a 4.1 kilobase (kb) segment at the insulin (INS) locus and their own data showing statistically significant associations between insulin-dependent diabetes mellitus (IDDM) and markers beyond the boundaries of that segment. We present data from an independent study of 201 IDDM patients and 107 non-diabetic control subjects that also show significant association with a marker 5′ of the INS locus. Patients and control subjects were genotyped at INS/+1140 A/C (a surrogate for the variable number tandem repeat (VNTR) polymorphism in the regulatory part of the INS gene) and a marker 5′ of the tyrosine hydroxylase (TH) gene, TH/pINS500-RsaI, making it 10 kb 5′ of the VNTR. Homozygotes for INS/+1140 allele ‘+’ were significantly more frequent among IDDM patients than among control subjects (73 vs 45%, p<0.001) giving an odds ratio of 3.3 (95% confidence interval (CI): 2.0–5.3). A very similar association was found for homozygotes for the TH/RsaIallele ‘+’ (53 vs 31%, p<0.001) giving an odds ratio of 2.6 (95% CI 1.6–4.2). By multilocus analysis, the TH/RsaI allele ‘+’ identified a subset of INS/+1140 alleles ‘+’ haplotypes that are more specifically associated with IDDM (odds ratio = 5.4, 95% CI 2.9–10.4) than allele +1140 ‘+’ as a whole. In conclusion, the segment of chromosome 11 that is associated with IDDM spans, at least, the INS and TH loci. No legitimate claim can be made that IDDM2 corresponds to the VNTR polymorphism at the INS locus until the correct boundaries for IDDM2 have been defined and other loci within them have been excluded as determinants of IDDM.
Similar content being viewed by others
Abbreviations
- IDDM:
-
Insulin-dependent diabetes mellitus
- INS :
-
insulin locus
- VNTR:
-
variable number of tandem repeats
- TH:
-
tyrosine hydroxylase
- IGF-2:
-
insulin-like growth factor-2
- DGGE:
-
denaturing gradient gel electrophoresis
References
Lucassen AM, Julier C, Beressi JP, Boitard C, Froguel P, Lathrop M, Bell JI (1993) Susceptibility to insulin dependent diabetes mellitus maps to a 4.1 kb segment of DNA spanning the insulin gene and associated VNTR. Nature Genet 4: 305–310
Bell GI, Karam JH, Rutter WJ (1981) Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc Natl Acad Sci USA 78: 5759–5763
Bell GI, Selby MJ, Rutter WJ (1982) The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature 295: 31–35
Bell GI, Horita S, Karam JH (1984) A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 33: 176–183
Rotwein P, Yokoyama S, Didier DK, Chirgwin JM (1986) Genetic analysis of the hypervariable region flanking the human insulin gene. Am J Hum Genet 39: 291–299
Bennett ST, Lucassen AM, Gough SCL et al. (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nature Genet 9: 284–292
Undlien DE, Bennett ST, Todd JA et al. (1995) Insulin gene region-encoded susceptibility to IDDM maps upstream of the insulin gene. Diabetes 44: 620–635
Barzilay J, Warram JH, Bak M, Laffel LMB, Canessa M, Krolewski AS (1992) Predisposition to hypertension: Risk factor for nephropathy and hypertension in IDDM. Kidney Inter 41: 723–730
Ulrich A, Dull TJ, Gray A, Brosius J, Sures I (1980) Genetic variation in the human insulin gene. Science 209: 612–615
O'Malley KL, Rotwein P (1988) Human tyrosine hydroxylase and insulin genes are contiguous on chromosome 11. Nucleic Acids Res 16: 4437–4446
Julier C, Hyer RN, Davies J et al. (1991) Insulin-IGF2 region on chromosome 11p encodes a gene implicated in HLA-DR4-dependent diabetes susceptibility. Nature 354: 155–159
Julier C, Lucassen A, Villedieu P et al. (1994) Multiple DNA variant association analysis: application to the insulin gene region in type 1 diabetes. Am J Hum Genet 55: 1247–1254
Parker S, Angelico MC, Laffel, L, Krolewski AS (1993) Application of denaturing gradient gel electrophoresis to detect DNA sequence differences encoding apolipoprotein E isoforms. Genomics 16: 245–247
Behrens J, Warram JH, Lerman LS, Krolewski AS (1990) User-friendly PC program to obtain DNA melting map. Am J Hum Genet 47 [Suppl I]:A173 (Abstract)
Fischer SG, Lerman LS (1983) DNA fragments differing by single base-pair substitution are separated in denaturing gradient gel: correspondence with melting theory. Proc Natl Acad Sci USA 80: 1579–1583
Cox NJ, Bell GI, Xiang K (1988) Linkage disequilibrium in the human insulin/insulin-like growth factor II region of human chromosome 11. Am J Hum Genet 43: 495–501
McGinnis RE, Spielman RS (1994) Linkage disequilibrium in the insulin gene region: size variation at the 5′ flanking polymorphism and bimodality among “Class I” alleles. Am J Hum Genet 55: 526–532
Rothman KJ (1986) Modern epidemiology. Little, Brown & Company, Boston, pp 85–114
Doria A, Warram JH, Rich SS, Krolewski AS (1994) Angiotensin I-converting enzyme (ACE): estimation of DNA haplotypes in unrelated individuals using denaturing gradient gel blots. Hum Genet 94: 117–123
McGinnis RE, Spielman RS (1995) Insulin expression: is VNTR allele 698 really anomalous? Nature Genet 10: 378–380
McGinnis RE, Spielman RS (1995) Insulin 5′ flanking polymorphism. Length of class 1 alleles in number of repeat units. Diabetes 44: 1296–1302
Catignani-Kennedy G, German MS, Rutter WJ (1995) The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nature Genet 9: 293–298
Lucassen AM, Screaton GR, Julier C, Elliot TJ, Lathrop M, Bell JI (1995) Regulation of insulin gene expression by the IDDM associated, insulin locus haplotype. Hum Mol Genet 4: 501–506
Jorde LB (1995) Linkage disequilibrium as a gene-mapping tool. Am J Hum Genet 56: 11–14
Akao Y, Utsumi KR, Naito K, Ueda R, Takahashi Y, Yamada K (1988) Gene encoding human p250 T-cell activation antigen maps to human chromosome 11. Somat Cell Molec Genet 14: 315–320
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Doria, A., Lee, J., Warram, J.H. et al. Diabetes susceptibility at IDDM2 cannot be positively mapped to the VNTR locus of the insulin gene. Diabetologia 39, 594–599 (1996). https://doi.org/10.1007/BF00403307
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00403307