Summary
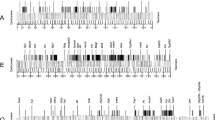
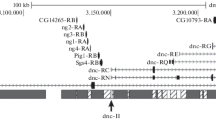
AT-rich segments were mapped in 6 different domains of the Drosophila melanogaster genome by partial denaturation of cloned genomic DNA segments and observation in the electron microscope (EM). As found in a previous investigation (Moreau et al., 1982), three types of AT-rich segments could be distinguished: 1) AT-rich linkers with a very high AT content, 600±200 bp long; 2) clusters of such AT-linkers extending over up to 10 Kpb and 3) AT-rich stretches which are shorter and of lower AT content compared to AT-linkers. Six genes previously localized in these domains were found to lie in relatively GC-rich segments framed by AT-rich elements of the 3 types: the Larval Serum Protein LSP-1 α gene is framed by 2 AT-linkers, the LSP-1β and LSP-2 genes by two AT-rich stretches, and the LSP-1γ gene by a cluster and a stretch. The 55 Kbp genomic segment encompassing the P6 gene contains and AT-cluster of about 15 Kbp and several AT-linkers and AT-rich stretches. The 85 Kbp domain containing the P1 gene includes 3 AT-rich clusters of about 10 Kbp each framing GC-rich domains punctuated by AT-linkers and stretches. This study shows that AT-mapping allows a rapid diagnosis of large genomic DNA domains in relation to the AT-rich segments which, possibly, are of significance with regard to genome organisation and function.
Similar content being viewed by others
References
Benton WD, Davis RW (1977) Screening gt recombinant clones by hybridization to single plaques in situ. Science 196:180–182
Blattner FR, Williams BG, Blechl AE, Denniston-Thompson K, Faber HE, Furlong LA, Grunwald DJ, Kieffer DO, Moore DD, Schumm JW, Sheldon EL, Smithies O (1977) Charon phages: safer derivatives of bacteriophage Lambda for DNA cloning. Science 196:161–169
Bridges L (1942) A new map of the salivary gland 3L chromosome. J Hered 33:403–408
Brock HW, Roberts DB (1980) Comparison of the larval serum proteins of Drosophila melanogaster using one and two-dimensional peptide mapping. Eur J Biochem 106:129–135
Case ST, Baker, RF (1975) Investigation into the use of Aspergillus oryzae S1 nuclease in the presence of solvents which destabilize or prevent DNA secondary structures: Formaldehyde, Formamide, and Glyoxal. Anal Biochem 64:477–484
Hall LMC, Mason PJ, Spierer P (1983) Transcripts, genes and bands in 315,000 Base-pairs of Drosophila DNA. J Mol Biol 169:83–96
Hayashi S, Gillam IC, Delaney AD, Tener GM (1978) Acetylation of chromosome squashes of Drosophila melanogaster decreases the background in autoradiographes from hybridization with I-labeled RNA. J Histochem Cytochem 26:677–679
Kejzlarova-Lepesant J, Brock HW, Moreau J, Dubertret M-L, Billault A, Lepesant J-A (1984) A complete and a truncated U1 SnRNA gene of Drosophila melanogaster are found as inverted repeat at region 82E of the polytene chromosomes. Nucl Acids Res 12:8835–8846
Lepesant JA, Levine M, Garen A, Lepesant-Kejzlarova J, Rat L, Somme-Martin G (1982) Developmentally regulated gene expression in Drosophila larval fat bodies. J Mol Appl Genetics 1:371–383
Lepesant JA, Maschat F, Kejzlarova-Lepesant J (1983) Ecdysteroid regulated gene expression in Drosophila melanogaster. The fat body system. Hormones and cell regulation. Vol 7, Dumont JE, Nunez J, Denton RM (eds) Elsevier/North Holland Biomedical Press, 255–268
McConaughy BL, Laird CD, McCarthy (1969) Nucleic acid reassociation in formamide. Biochemistry 8:3289–3295
Maniatis T, Hardison RC, Lacy E, Lauer J, O'Connell C, Quon D, Sim GK, Efstratiadis A (1978) The isolation of structural genes from librairies of Eukaryotic DNA. Cell 15:687–701
Moreau J, Matyash-Smirniaguina L, Scherrer K (1981) Systematic punctuation of eukaryotic DNA by A+T-rich sequences. Proc Natl Acad Sci USA 78:1341–1345
Moreau J, Marcaud L, Maschat F, Kejzlarova-Lepesant J, Lepesant JA, Scherrer K (1982) A+T-rich linkers define functional domains in eukaryotic DNA. Nature 295:260–262
Mount S, Steitz J (1981) Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucl Acids Res 9:6351–6368
Rigby PWJ, Dieckmann M, Rhodes C, Berg P (1977) Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol 113:237–251
Scherrer K (1971) Adenosine-rich sequences in rapidly hybridizing messenger like RNA and their possible significance for reiterated base sequences in Eukaryotic DNA. FEBS Letter 17:68–72
Scherrer K (1974) Control of gene expression in animal cells: the cascade regulation hypothesis revisited. Kohn, Shatkay, (eds) Adv Expl Med and Biol, Vol 44, pp 169–219, Plenum Press, New York
Schildkraut C, Lifson S (1965) Dependence of the melting temperature of DNA on salt concentration. Biopolymer. 3:195–208
Smith DF, McClelland A, White BN, Addison CF, Glover DM (1981) The molecular cloning of a dispersed set of developmentally regulated genes which encode the major larval serum protein of Drosophila melanogaster. Cell 23:441–449
Spierer P, Spierer A, Bender W, Hogness DS (1983) Molecular mapping of genetic and chromomeric units in Drosophila melanogaster. J Mol Biol 168:35–50
Strauss F, Gaillard C, Prunell A (1981) Helical periodicity of DNA, poly(dA). Poly(dT) and poly(dA-dT). Poly(dA-dT) in solution. Eur J Biochem 118:215–222
Strobel E, Dunsmuir P, Rubin GM (1979) Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell 17:429–439
Tibbetts C, Johansson K, Philipson L (1973) Hydroxyapatite chromatography and formamide denaturation and adenovirus DNA. J Virol 12:218–225
Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G (1970) Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40:734–744
Yedvobnick B, Levine M (1982) The LSP-2 gene and a 5′ flanking sequence are independently expressed in Drosophila melanogaster. Nature 297:239–241
Author information
Authors and Affiliations
Additional information
Communicated by E.K.F. Bautz
Rights and permissions
About this article
Cite this article
Moreau, J., Kejzlarovä-Lepesant, J., Brock, H. et al. AT-rich linkers in long range organisation of Drosophila DNA. Mol Gen Genet 199, 357–364 (1985). https://doi.org/10.1007/BF00330743
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00330743