Abstract
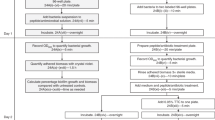
Many bacteriologic studies, including cellular invasion and adherence assays, require enumeration of viable organisms or colony forming units. When attempting to screen large numbers of clinical or environmental isolates or laboratory-derived mutants for differences in invasion or adherence phenotype, standard plating methods can be cumbersome and severely limit the number of organisms or conditions which can be tested. As a potential alternative, we describe a simple, rapid and inexpensive soft agar-based technique for semi-quantitative determination of bacterial colony counts directly within the wells of a 96-well microtiter plate.
Similar content being viewed by others
References
Amann RI, Zarda B, Stahl DA, et al. (1992). Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 58: 3007–3011.
Devenish JA, Schiemann DA (1981). HeLa cell infection by Yersinia enterocolitica: evidence for lack of intracellular multiplication and development of a new procedure for quantitative expression of infectivity. Infect Immun 32: 48–55.
Francisco DE, Mah RA, Rabin AC (1973). Acridine orange epifluorescent technique for counting bacteria in natural waters. Trans Am Microsc Soc 92: 416–421.
Lundgren B (1981). Fluorescein diacetate as a stain of metabolically active bacteria in soil. Oikos 36: 17–22.
Noble PA, Ashton E, Davidson CA, et al. (1991). Heterotrophic plate counts of surface water samples by using impedance methods. Appl Environ Microbiol 57: 3287–3291.
Norris KP, Powell EO (1961). Improvements in determining total counts of bacteria. J R Micros Soc 80: 107–119.
Peck R (1985). A one-plate assay for macrophage bactericidal activity. J Immunol Methods 82: 131–140.
Pinder AC, Purdy PW, Poulter SA, et al. (1990). Validation of flow cytometry for rapid enumeration of bacterial concentrations in pure cultures. J Appl Bacteriol 69: 92–100.
Rubens CE, Smith S, Hulse M, et al. (1992). Respiratory epithelial cell invasion by group B streptococci. Infect Immun 60: 5157–5163.
Selan LE, Berlutti FN, Passariello CE, Thaller MC, Renzini G (1992). Reliability of a bioluminiscence ATP assay for detection of bacteria. J Clin Microbiol 30: 1739–1742.
Sullam PM, Bayer AS, Foss WM, et al. (1996). Diminished platelet binding in vitro by Staphylococcus aureus is associated with reduced virulence in a rabbit model of infective endocarditis. Infect Immun 64: 4915–4921.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nizet, V., Smith, A.L., Sullam, P.M. et al. A simple microtiter plate screening assay for bacterial invasion or adherence. Methods Cell Sci 20, 107–111 (1998). https://doi.org/10.1023/A:1009830606819
Issue Date:
DOI: https://doi.org/10.1023/A:1009830606819