Abstract
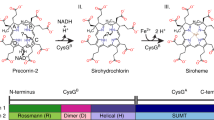
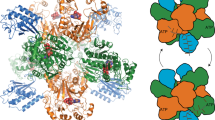
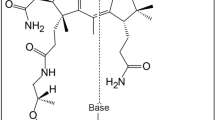
Sulfur metabolism depends on the iron-containing porphinoid siroheme. In Salmonella enterica, the S-adenosyl-L-methionine (SAM)-dependent bismethyltransferase, dehydrogenase and ferrochelatase, CysG, synthesizes siroheme from uroporphyrinogen III (uro'gen III). The reactions mediated by CysG encompass two branchpoint intermediates in tetrapyrrole biosynthesis, diverting flux first from protoporphyrin IX biosynthesis and then from cobalamin (vitamin B12) biosynthesis. We determined the first structure of this multifunctional siroheme synthase by X-ray crystallography. CysG is a homodimeric gene fusion product containing two structurally independent modules: a bismethyltransferase and a dual-function dehydrogenase-chelatase. The methyltransferase active site is a deep groove with a hydrophobic patch surrounded by hydrogen bond donors. This asymmetric arrangement of amino acids may be important in directing substrate binding. Notably, our structure shows that CysG is a phosphoprotein. From mutational analysis of the post-translationally modified serine, we suggest a conserved role for phosphorylation in inhibiting dehydrogenase activity and modulating metabolic flux between siroheme and cobalamin pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Murphy, M.J., Siegel, L.M., Tove, S.R. & Kamin, H. Siroheme: a new prosthetic group participating in six-electron reduction reactions catalyzed by both sulfite and nitrite reductases. Proc. Natl. Acad. Sci. USA 71, 612–616 (1974).
Murphy, M.J. & Siegel, L.M. Siroheme and sirohydrochlorin. The basis for a new type of porphyrin-related prosthetic group common to both assimilatory and dissimilatory sulfite reductases. J. Biol. Chem. 248, 6911–6919 (1973).
Siegel, L.M., Murphy, M.J. & Kamin, H. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of enterobacteria. I. The Escherichia coli hemoflavoprotein: molecular parameters and prosthetic groups. J. Biol. Chem. 248, 251–264 (1973).
Christner, J.A., Munck, E., Janick, P.A. & Siegel, L.M. Mössbauer spectroscopic studies of Escherichia coli sulfite reductase. Evidence for coupling between the siroheme and Fe4S4 cluster prosthetic groups. J. Biol. Chem. 256, 2098–2101 (1981).
Janick, P.A. & Siegel, L.M. Electron paramagnetic resonance and optical spectroscopic evidence for interaction between siroheme and Fe4S4 prosthetic groups in Escherichia coli sulfite reductase hemoprotein subunit. Biochemistry 21, 3538–3547 (1982).
Crane, B.R., Siegel, L.M. & Getzoff, E.D. Sulfite reductase structure at 1.6 Å: evolution and catalysis for reduction of inorganic anions. Science 270, 59–67 (1995).
Siegel, L.M. Biochemistry of the Sulfur Cycle (Academic, New York, 1975).
Peck, H.D. & Lissolo, T. The Nitrogen and Sulfur Cycles 99–132 (Cambridge Univ. Press, Cambridge, 1988).
Scott, A.I. How nature synthesizes vitamin B12—a survey of the last four billion years. Angew. Chem. Int. Edn. Engl. 32, 1223–1376 (1993).
Jordan, P.M. Highlights in haem biosynthesis. Curr. Opin. Struct. Biol. 4, 902–911 (1994).
Blanche, F., Debussche, L., Thibaut, D., Crouzet, J. & Cameron, B. Purification and characterization of S-adenosyl-L-methionine: uroporphyrinogen III methyltransferase from Pseudomonas denitrificans. J. Bacteriol. 171, 4222–4231 (1989).
Raux, E., Schubert, H.L. & Warren, M.J. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell Mol. Life Sci. 57, 1880–1893 (2000).
Warren, M.J. et al. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem. J. 302, 837–844 (1994).
Warren, M.J. et al. Enzymatic synthesis of dihydrosirohydrochlorin (precorrin-2) and of a novel pyrrocorphin by uroporphyrinogen III methylase. FEBS Lett. 261, 76–80 (1990).
Jeter, R.M., Olivera, B.M. & Roth, J.R. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J. Bacteriol. 159, 206–213 (1984).
Raux, E. et al. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem. J. 370, 505–516 (2003).
Raux, E., McVeigh, T., Peters, S.E., Leustek, T. & Warren, M.J. The role of Saccharomyces cerevisiae Met1p and Met8p in sirohaem and cobalamin biosynthesis. Biochem. J. 338, 701–708 (1999).
Schubert, H.L. et al. The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase. EMBO J. 21, 2068–2075 (2002).
Schubert, H.L., Wilson, K.S., Raux, E., Woodcock, S.C. & Warren, M.J. The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase. Nat. Struct. Biol. 5, 585–592 (1998).
Crouzet, J. et al. Genetic and sequence analysis of an 8.7-kilobase Pseudomonas denitrificans fragment carrying eight genes involved in transformation of precorrin-2 to cobyrinic acid. J. Bacteriol. 172, 5980–5990 (1990).
Keller, J.P. et al. The crystal structure of MT0146/CbiT suggests that the putative precorrin-8w decarboxylase is a methyltransferase. Structure 10, 1475–1487 (2002).
Goldman, B.S. & Roth, J.R. Genetic structure and regulation of the cysG gene in Salmonella typhimurium. J. Bacteriol. 175, 1457–1466 (1993).
Raux, E., Thermes, C., Heathcote, P., Rambach, A. & Warren, M.J. A role for Salmonella typhimurium cbiK in cobalamin (vitamin B12) and siroheme biosynthesis. J. Bacteriol. 179, 3202–3212 (1997).
Schubert, H.L., Raux, E., Wilson, K.S. & Warren, M.J. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry 38, 10660–10669 (1999).
Fazzio, T.G. & Roth, J.R. Evidence that the CysG protein catalyzes the first reaction specific to B12 synthesis in Salmonella typhimurium, insertion of cobalt. J. Bacteriol. 178, 6952–6959 (1996).
Belyaeva, T., Griffiths, L., Minchin, S., Cole, J. & Busby, S. The Escherichia coli cysG promoter belongs to the 'extended −10' class of bacterial promoters. Biochem. J. 296, 851–857 (1993).
Burns, H.D., Belyaeva, T.A., Busby, S.J. & Minchin, S.D. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem. J. 317, 305–311 (1996).
Robin, C. et al. Primary structure, expression in Escherichia coli, and properties of S-adenosyl-L-methionine:uroporphyrinogen III methyltransferase from Bacillus megaterium. J. Bacteriol. 173, 4893–4896 (1991).
Mascaro, L. Jr. et al. Synthesis of methionine carrying a chiral methyl group and its use in determining the steric course of the enzymatic C-methylation of indolepyruvate during indolmycin biosynthesis. J. Am. Chem. Soc. 99, 273–274 (1977).
Warren, M.J., Gonzalez, M.D., Williams, H.J., Stolowich, N.J. & Scott, A.I. Uroporphyrinogen III methylase catalyzes the enzymatic synthesis of sirohydrochlorins II and IV by a clockwise mechanism. J. Am. Chem. Soc. 112, 5343–5345 (1990).
Eschenmoser, A. Vitamin B12: experiments concerning the origin of its molecular structure. Angew. Chem. Int. Edn. Engl. 27, 5–39 (1988).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
Griffiths, L. & Cole, J.A. Lack of redox control of the anaerobically-induced nirB+ gene of Escherichia coli K-12. Arch. Microbiol. 147, 364–369 (1987).
Rossmann, M.G. Comparison of the evolution of heme binding and NAD binding proteins. Fed. Proc. 35, 2112–2114 (1976).
Woodcock, S.C. et al. Effect of mutations in the transmethylase and dehydrogenase/chelatase domains of sirohaem synthase (CysG) on sirohaem and cobalamin biosynthesis. Biochem. J. 330, 121–129 (1998).
Woodcock, S.C. & Warren, M.J. Evidence for a covalent intermediate in the S-adenosyl-L-methionine–dependent transmethylation reaction catalysed by sirohaem synthase. Biochem. J. 313, 415–421 (1996).
Warren, M.J., Bolt, E. & Woodcock, S.C. 5-Aminolaevulinic acid synthase and uroporphyrinogen methylase: two key control enzymes of tetrapyrrole biosynthesis and modification. Ciba Found. Symp. 180, 26–40 (1994).
Roessner, C.A. Use of cobA and cysGA as red fluorescent indicators. Methods Mol. Biol. 183, 19–30 (2002).
Dean, A.M. & Koshland, D.E. Jr. Electrostatic and steric contributions to regulation at the active site of isocitrate dehydrogenase. Science 249, 1044–1046 (1990).
Hurley, J.H., Dean, A.M., Sohl, J.L., Koshland, D.E. Jr. & Stroud, R.M. Regulation of an enzyme by phosphorylation at the active site. Science 249, 1012–1016 (1990).
Stryer, L. Biochemistry 524 (Freeman, New York, 1998).
Fujino, E., Fujino, T., Karita, S., Sakka, K. & Ohmiya, K. Cloning and sequencing of some genes responsible for porphyrin biosynthesis from the anaerobic bacterium Clostridium josui. J. Bacteriol. 177, 5169–5175 (1995).
Roessner, C.A., Huang, K.X., Warren, M.J., Raux, E. & Scott, A.I. Isolation and characterization of 14 additional genes specifying the anaerobic biosynthesis of cobalamin (vitamin B12) in Propionibacterium freudenreichii (P. shermanii). Microbiology 148, 1845–1853 (2002).
Wu, J.Y., Siegel, L.M. & Kredich, N.M. High-level expression of Escherichia coli NADPH-sulfite reductase: requirement for a cloned cysG plasmid to overcome limiting siroheme cofactor. J. Bacteriol. 173, 325–333 (1991).
de La Fortelle, E. & Bricogne, G. Maximum-Likelihood Heavy-Atom Parameter Refinement for the Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods 472–494 (Academic, New York, 1997).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Abrahams, J.P. & Leslie, A.W.G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996).
McRee, D.E. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
M.E.S. was supported by a predoctoral US National Science Foundation fellowship and the Skaggs Institute for Chemical Biology. This research was funded by a grant from the US National Institutes of Health (NIH) to E.D.G. M.J.W. acknowledges funding from the UK Biotechnology and Biological Research Council and the Wellcome Trust. We thank D. Barondeau, M. Didonato, J. Huffman and C. Kassmann for useful discussions, A. Arvai and E. Garcin for assistance with data collection and M. Pique for help generating Figure 6b,c. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy (DOE), Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DOE, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Stroupe, M., Leech, H., Daniels, D. et al. CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat Struct Mol Biol 10, 1064–1073 (2003). https://doi.org/10.1038/nsb1007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb1007
This article is cited by
-
Siroheme synthase orients substrates for dehydrogenase and chelatase activities in a common active site
Nature Communications (2020)
-
dissectHMMER: a HMMER-based score dissection framework that statistically evaluates fold-critical sequence segments for domain fold similarity
Biology Direct (2015)
-
The sulfur/sulfonates transport systems in Xanthomonas citri pv. citri
BMC Genomics (2015)
-
Genome-scale analysis reveals a role for NdgR in the thiol oxidative stress response in Streptomyces coelicolor
BMC Genomics (2015)
-
Recent advances in the biosynthesis of modified tetrapyrroles: the discovery of an alternative pathway for the formation of heme and heme d 1
Cellular and Molecular Life Sciences (2014)