Summary
Isozyme analysis was used to study the genetic variation and the genetic relationships of a collection of 48 Brassica spp. including 31 Portuguese coles (Brassica oleracea L.) accessions representative of the different landraces cultivated in Portugal. Other brassicas included in this experiment were Jersey kale, kailaan, common cabbages, broccolis, cauliflower, nine-chromosome wild brassica and turnip. Nine enzymes used in the starch gel electrophoresis included: PGM, PGI, AAT, LAP, TPI, FBP, SOD, IDH and GR. Twenty-one putative loci were revealed, with 3 showing invariance and the other 18 contained 50 alleles. The allelic frequencies at these loci represented by 40 plants per accession were used to calculate the following estimators of genetic variation: % of polymorphic loci, average number of alleles per polymorphic loci, average number of alleles per locus, and index of heterozygosity. The genetic relationships were evaluated considering Nei (1978) and Rogers (1972) genetic distances between each pair of accessions whose matrices were hierarchically clustered by the UPGMA method. The accessions were also studied using the principal coordinate analysis.
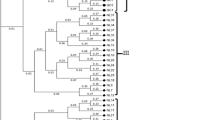
Portuguese Tronchuda cabbages and Galega kales have shown high genetic diversity in comparison with the other accessions. This indicates their potential variation for use in breeding programs. The UPGMA results show that the 48 accessions, with the exception of B. insularis, B. cretica, and turnip, can be clustered into 6 groups: (a) Portuguese Tronchuda cabbages, Galega kales and Algarve cabbages; (b) common cabbages and kales; (c) Couve Poda do Algarve and broccoli; (d) Algarve cabbage and common cabbages; (e) kailaan; (f) broccoli and cauliflower. The groupings obtained by the isozyme analysis are difficult to interpret considering the origin of the Brassica spp. and the morphological resemblance among the accessions.
Similar content being viewed by others
References
Allen, B.W., P.W.Goodenough, J.S.C.Lee & P.P.Rutherford, 1986. Cauliflower curds of different cultivars show distinct isoenzyme differences in acid phosphatases and aspartate aminotransferase. Trans. Biochem. Soc. London 14: 455–456.
Arús, P. & T.J.Orton, 1983. Inheritance and linkage relationships of isozyme loci in Brassica oleracea. J. Hered. 74: 405–412.
Brown, A.H.D., 1979. Enzyme polymorphism in plant populations. Theor. Pop. Biol. 15: 1–24.
Brown, A.H.D. & B.S.Weir, 1983. Measuring genetic variability in plant populations. In: S.D.Tanksley & T.J.Orton (Eds). Isozymes in Plant Genetics and Breeding. Part A. Elsevier Science Publishers B.V., Amsterdam. pp. 219–239.
Chèvre, A.M., R.Delourme, F.Eber, P.Arús & C.F.Quiros, 1990. Allele nomenclature for five isozyme systems in Brassica nigra, Brassica campestris and Brassica oleracea. Proc. 6th Crucifer Genetics Workshop. Cornell Univ., Ithaca, New York. pp. 19–20. (Abstract).
Coulthart, M.B., 1979. A Comparative Electrophoretic Study of an Allopolyploid Complex in Brassica. M.Sc. Thesis, University of Alberta, Canada. 114 pp.
Crisp, P., 1982. The use of an evolutionary scheme for cauliflowers in the screening of genetic resources. Euphytica 31: 725–734.
Dias, J.S., M.B.Lima, K.M.Song, A.A.Monteiro, P.W.Williams & T.C.Osborn, 1992. Molecular taxonomy of Portuguese Tronchuda cabbage and kale landraces using nuclear RFLPs. Euphytica 58: 221–229.
Dias, J.S., M.E.Ferreira & P.W.Williams, 1993a. Screening of Portuguese cole landraces (Brassica oleracea L.) with Peronospora parasitica and Plasmodiophora brassicae. Euphytica 67: 135–141.
Dias, J.S., A.A.Monteiro & M.B.Lima, 1993b. Numerical taxonomy of Portuguese Tronchuda cabbage and Galega kale landraces using morphological characters. Euphytica 69: 51–68.
Ferreira, M.E., J.S.Dias, A.Mengistu & P.W.Williams, 1993. Screening of Portuguese cole landraces (Brassica oleracea L.) with Leptosphaeria maculans and Xanthomonas campestris pv. campestris. Euphytica 65: 219–227.
Gottlieb, L.D., 1981. Electrophoretic evidence and plant populations. Progress in Phytochemistry 7: 1–46.
Gower, J.C., 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 27: 325–338.
Kuman, R. & V.P.Gupta, 1985. Isozyme studies in Indian mustard (Brassica juncea L.). Theor. Appl. Genet. 70: 107–110.
May, B., J.E.Marsden & C.G.Shenck, 1988. Electrophoretic Procedures, Recipes and Nomenclature Used in Cornell Laboratory for Ecological and Evolutionary Genetics. Cornell Univ. New York. 20 pp. (Ciclost.).
Monteiro, A.A. & P.W.Williams, 1989. The exploration of genetic resources of Portuguese cabbage and kale for resistance to several Brassica diseases. Euphytica 41: 215–225.
Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Quiros, C.F., 1987. Duplicated isozyme loci and cellular compartmentation of their products in Brassica. Cruciferae Newsletter 12: 24.
Rogers, I.S., 1972. Measures of genetic similarity and genetic distance. Univ. Texas Publ. 7213: 145–153.
Rohlf, F.J., 1989. NTSYS-pc. Numerical Taxonomy and Multivariate Analysis System. Version 1.5. Exeter Publishing Ltd. Setauket, New York, USA.
Thorpe, M.L., L.H.Duke & W.D.Beversdorf, 1987. Procedures for the Detection of Isozymes of Rapeseed (Brassica napus and Brassica campestris) by Starch Gel Electrophoresis. Tech. Bull. OAC 887. Dept. Crop Sci. Univ. Guelph, Canada. 66 pp.
Vaughan, J.G., A.Waite, D.Boulter & S.Waiters, 1966. Comparative studies of the seed proteins of Brassica campestris, Brassica oleracea and Brassica nigra. J. Exp. Bot. 17: 332–343.
Weeden, N.F., 1982. Starch Gel Isozyme Electrophoresis Techniques Used in Genetic and Taxonomic Studies with Bean, Pea, Squash, Cabbage, and Lettuce. Genetic Resources Unit and Tropical Pastures Program. CIAT, Cali, Colombia. (Compiled by Hernando Ramirez.) 42 pp. (Ciclost.)
Wendel, J.F. & N.F.Weeden, 1989. Visualization and interpretation of plant isozymes. In: D.E.Soltis & P.S.Soltis (Eds). Isozymes in Plant Biology. Dioscorides Press, Washington. pp. 5–43.
Williams, P.H. & C.B.Hill, 1986. Rapid cycling populations of Brassica. Science 232: 1385–1389.
Yavada, J.S., J.B.Chowdhury, S.N.Kakar & H.S.Nainawatee, 1979. Comparative electrophoretic studies of proteins and enzymes of some Brassica species. Theor. Appl. Genet. 54: 89–91.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Dias, J.S., Monteiro, A.A. & Kresovich, S. Genetic diversity and taxonomy of Portuguese Tronchuda cabbage and Galega kale landraces using isozyme analysis. Euphytica 75, 221–230 (1994). https://doi.org/10.1007/BF00025607
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00025607