Abstract
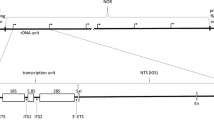
We have used nascent strand determination analysis to map start sites of DNA replication in the mouse ribosomal gene cluster in which individual copies of the ribosomal genes are separated by intergenic spacer regions. One origin of bidirectional replication (OBR) was localized within a 3 kb region centered about 1.6 kb apstream of the rDNA transcription start site. At least one additional initiation site is situated near the 3′ end of the transcription unit. Adjacent to the OBR at the transcription start site are located two amplification-promoting sequences, i.e., APS1 and APS2. Nuclease-hypersensitive sites were identified in both the two APSs as well as in the OBR region, thus indicating that these sequences have an altered chromatin structure. In the OBR an intrinsically bent region, a purine-rich element and other prospective initiation zone components are found.
Similar content being viewed by others
References
AnachkovaB, HamlinJL (1989) Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol 9: 532–540
BenardM, LagnelC, PierronG (1995) Site-specific initiation of DNA replication within the non-transcribed spacer ofPhysarum rDNA. Nucleic Acids Res 23: 1447–1453
BerberichS, LeffakM (1993) DNase-sensitive chromatin structure near a chromosomal origin of bidirectional replication of the avian α-globin locus. DNA Cell Biol 12: 703–714
BergemanAD, JohnsonEM (1992) The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol 12: 1257–1265
BotchanPM, DaytonAI (1982) A specific replication origin in the chromosomal rDNA ofLytechinus variegatus. Nature 299: 453–456
BozzoniI, BaldariCT, AmaldiR, Buongiorno-NardelliM (1981) Replication of ribosomal DNA inXenopus laevis. Eur J Biochem 118: 585–590
BraatenDC, ThomasJR, LittleRD, DicksonKR, GoldbergI, SchlessingerD, CiccodicolaA, D'UrsoM (1988) Locations and contexts of sequences that hybridize to poly (dG-dT)·(dC-dA) in mammalian ribosomal DNAs and two X-linked genes. Nucleic Acids Res 16: 865–881
BrewerBJ, FangmanWL (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643
BurhansWC, SelequeJE, HeintzNH (1986) Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci USA 83: 7790–7794
BurhansWC, VassilievLT, CaddleMS, HeintzNH, DePamphilisML (1990) Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell 62: 955–965
CaddleMS, LussierRH, HeintzNH (1990) Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol 211: 19–23
CoffmanFD, GeorgoffI, FresaKL, SylvesterJ, GonzalezI, CohenS (1993) In vitro replication of plasmids containing human ribosomal gene sequences: origin localization and dependence of an aprotinin-binding cytosolic protein. Exp Cell Res 209: 123–132
CoverleyD, LaskeyRA (1994) Regulation of eukaryotic DNA replication. Annu Rev Biochem 63: 745–776
DePamphilisML (1988) Transcriptional elements as components of eukaryotic origins of DNA replication. Cell 52: 635–638
DePamphilisML (1993a) Origins of DNA replication that function in eukaryotic cells. Curr Opin Cell Biol 5: 434–441
DePamphilisML (1993b) Origins of DNA replication in metazoan chromosomes. J Biol Chem 268: 1–4
DePamphilisML (1993c) Eukaryotic DNA replication: Anatomy of an origin. Annu Rev Biochem 62: 29–63
DijkwelPA, HamlinJL (1992) Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid Mimosine. Mol Cell Biol 12: 3715–3722
EckdahlTT, AndersonJN (1990) Conserved DNA structures in origins of replication. Nucleic Acids Res 18: 1609–1612
FangmanWL, BrewerBJ (1991) Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol 7: 375–402
HandeliS, KlarM, MeuthM, CedarH (1989) Maping replication units in animal cells. Cell 57: 909–920
HerbomelP, SaragostiS, BlangyD, YanivM (1981) Fine structure of the origin-proximal DNAseI-hypersensitive region in wild-type and EC mutant polyoma. Cell 25: 651–658
HernandezPL, BjerknesCA, LammSS, Van'tHofJ (1988) Proximity of an ARS consensus sequence to a replication origin of pea (Pisum sativum). Plant Mol Biol 10: 413–422
KooHS, WuHM, CrothersDM (1986) DNA bending at adenine thymine tracts. Nature 320: 501–506
KuhnA, DeppertA, GrummtI (1990) A 140-base-pair repetitive sequence element in the mouse rRNA gene spacer enhances transcription by RNA polymerase I in a cell-free system. Proc Natl Acad Sci USA 87: 7527–7531
LinskensMHK, HubermanJA (1988) Organization of replication of ribosomal DNA inSaccharomyces cerevisiae. Mol Cell Biol 8: 4927–4935
LittleRD, PlattTHK, SchildkrautCL (1993) Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol 13: 6600–6613
PalenEG, CechTR (1984) Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes ofTetrahymena. Cell 36: 933–941
PanWJ, GallagherRC, BlackburnEH (1995) Replication of an rRNA gene origin plasmid. in theTetrahymena thermophila macronucleus is prevented by transcription through the origin from an RNA polymerase I promoter. Mol Cell Biol 15: 3372–3381
PutnamCD, CopenhaverGP, DentonML, PikaardCS (1994) The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol Cell Biol 14: 6476–6488
ReevesR (1992) Chromatin changes during the cell cycle. Curr Opin Cell Biol 4: 313–423
SafferLD, MillerOL (1986) Electron microscopic study ofSaccharomyces cerevisiae rDNA chromatin replication. Mol Cell Biol 6: 1148–1157
SaragostiS, MoyneG, YanivM (1980) Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell 20: 65–73
StolzenburgF, GerwigR, DinklE, GrummtF (1994) Structural homologies and functional similarities between mammalian origins of replication and amplification promoting sequences. Chromosoma 103: 209–214
StraussF, VarshavskyAJ (1984) A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell 37: 889–901
TruettMA, GallJG (1977) The replication of ribosomal DNA in the macronucleus of Tetrahymena. Chromosoma 64: 295–303
Van'tHofJ, LammSS (1992) Site of initiation of replication of the ribosomal genes of pea (Pisum sativum) detected by two-dimensional gel electrophoresis. Plant Mol Biol 20: 377–382
VarshavskyAJ, SundinOH, BohnMJ (1979) A stretch of “late” SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell 16: 453–466
VassilevL, JohnsonEM (1989) Mapping initiation sites of DNA replicationin vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res 17: 7693–7705
VassilevL, JohnsonEM (1990) An initiation zone of chromosomal DNA replication located upstream of thec-myc gene in proliferating HeLa cells. Mol Cell Biol 10: 4899–4904
VassilevLT, BurhansWC, DePamphilisML (1990) Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol 10: 4685–4689
VogtVM, BraunR (1977) The replication of ribosomal DNA inPhysarum polycephalum. Eur J Biochem 80: 557–566
WegnerM, ZastrowG, KlaviniusA, SchwenderS, MüllerF, LukszaH, HoppeJ, WienbergJ, GrummtF (1989)Cis-acting sequences from mouse rDNA promote plasmid DNA amplification and persistence in mouse cells: implication of HMG-I in their function. Nucleic Acids Res 17: 9909–9932
WegnerM, SchwenderS, DinklE, GrummtF (1990) Interaction of a protein with a palindromic sequence from murine rDNA increases the occurrence of amplification-dependent transformation in mouse cells. J Biol Chem 265: 13925–13932
WuHM, CrothersDM (1984) The locus of sequence-directed and protein-induced DNA bending. Nature 308: 509–513
YoonY, SanchezJA, BrunC, HubermanJA (1995) Mapping of replication initiation sites in human ribosomal DNA by nascent-strand abundance analysis. Mol Cell Biol 15: 2482–2489
ZhaoK, KäsE, GonzalezE, LaemmliK (1993) SAR-dependent mobilization of histone H1 by HMG-I/Yin vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J 12: 3237–3247
ZwiebC, KimJ, AdhyaS (1989) DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev 3: 606–611
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gögel, E., Längst, G., Grummt, I. et al. Mapping of replication initiation sites in the mouse ribosomal gene cluster. Chromosoma 104, 511–518 (1996). https://doi.org/10.1007/BF00352115
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00352115