Abstract
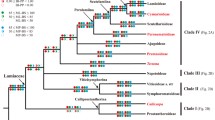
Thirty-two partial phytochrome sequences from algae, mosses, ferns, gymnosperms, and angiosperms (11 of them newly released ones from our laboratory) were analyzed by distance and characterstate approaches (PHYLIP, TREECON, PAUP). In addition, 12 full-length sequences were analyzed. Despite low bootstrap values at individual internal nodes, the inferred trees (neighbor joining, Fitch, maximum parsimony) generally showed similar branching orders consistent with other molecular data. Lower plants formed two distinct groups. One basal group consisted ofSelaginella, Equisetum, and mosses; the other consisted of a monophyletic cluster of frond-bearing pteridophytes.Psilotum was a member of the latter group and hence perhaps was not, as sometimes suggested, a close relative of the first vascular plants. The results further suggest that phytochrome gene duplication giving rise to a- and b- and later to c-types may have taken place within seedfern genomes. Distance matrices dated the separation of mono- and dicotyledons back to about 260 million years before the present (Myrb.p.) and the separation ofMetasequoia andPicea to a fossil record-compatible value of 230 Myr B.P. TheEphedra sequence clustered with the c- or a-type andMetasequoia andPicea sequences clustered with the b-type lineage. The “paleoherb”Nymphaea branched off from the c-type lineage prior to the divergence of mono- and dicotyledons on the a- and b-type branches. Sequences ofPiper (another “paleoherb”) created problems in that they branched off from different phytochrome lineages at nodes contradicting distance from the inferred trees' origin.
Similar content being viewed by others
References
Adam E, Deak M, Kay S, Chua N-H, Nagy F (1993) Sequence of a tobacco (Nicotiana tabacum) gene coding for type A phytochrome. Plant Physiol 101:1407–1408
Bernardi G, Mouchiroud D, Gautier C (1993) Silent substitutions in mammalian genomes and their evolutionary implications. J Mol Evol 37:583–589
Christensen AH, Quail PH (1989) Structure and expression of a maize phytochrome encoding gene. Gene 85:381–390
Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family inArabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mot Biol 25:413–427
Collins DW, Jukes TH (1993) Relationship between G + C in silent sites of codons and amino acid composition of human proteins. J Mol Evol 36:201–213
Dehesh K, Tepperman J, Christensen AH, Quail PH (1991) phyB is evolutionarily conserved and constitutively expressed in rice seedling shoots. Mol Gen Genet 225:305–313
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Feng DF, Doolittle RF (1990) Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol 183:375–387
Gaut BS, Muse SV, Clark WD, Clegg MT (1992) Relative rates of nucleotide substitutions of the rbcL locus of monocotyledonous plants. J Mol Evol 35:292–303
Halliday KJ, Koornneef M, Whitelam GC (1994) Phytochrome B and at least one other phytochrome mediate the accelerated flowering response ofArabidopsis thaliana L. to low-red/far-red ratio. Plant Physiol 104:1311–1315
Hamby RK, Zimmer EA (1992) Ribosomal RNA as a phylogenetic tool in plant systematics. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Routledge Chapman Hall, London, pp 50–91
Hanelt S, Braun B, Marx S, Schneider-Poetsch HAW (1992) Phytochrome evolution: a phylogenetic tree with the first complete sequence of a phytochrome from a cryptogamic plant (Selaginella martensii Spring). Photochem Photobiol 56:751–758
Hasebe M, Ito M, Kofuji R, Ueda K, Iwatsuki K (1993) Phylogenetic relationships of ferns deduced from rbcL gene sequence. J Mol Evol 37:476–482
Hershey HP, Barker RF, Idler KB, Murray MG, Quail PH (1987) Nucleotide sequence and characterization of a gene encoding the phytochrome polypeptide fromAvena. Gene 61:339–348
Heyer A, Gatz C (1992a) Isolation and characterization of a cDNAclone coding for potato type A phytochrome. Plant Mol Biol 18:535–544
Heyer A, Gatz C (1992b) Isolation and characterization of a cDNAclone coding for potato type B phytochrome. Plant Mol Biol 20: 589–600
Hiesel R, von Haeseler A, Brennicke A (1994) Plant mitochondrial nucleic acid sequences as a tool for phylogenetic analysis. Proc Natl Acad Sci USA 91:634–638
Kay SA, Keith B, Shinozaki K, Chye ML, Chua N-H (1989) The sequence of the rice phytochrome gene. Nucleic Acids Res 17: 2865–2866
Kendrick RE, Kronenberg GHM (eds) (1994) Photomorphogenesis in plants, 2nd. ed. Kluwer, Dordrecht
Kern R, Gasch A, Deak M, Kay SA, Chua N-H (1993) phyB of tobacco, a new member of the phytochrome family. Plant Physiol 102:1363–1364
Kolukisaoglu HÜ, Braun B, Martin WF, Schneider-Poetsch HAW (1993) Mosses do express conventional, distantly B-type-related phytochromes: phytochrome ofPhyscomitrella patens (Hedw.). FEBS Lett 334:95–100
Kwiatowski J, Skarecky D, Bailey K, Ayala FJ (1994) Phylogeny ofDrosophila and related genera inferred from the nucleotide sequence of the Cu,Zn Sod gene. J Mol Evol 38:443–454
Li W-H (1993) Unbiased estimation of rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99
Lopez-Figueroa F, Lindemann P, Braslavsky SE, Schaffner K, Schneider-Poetsch HAW, Rüdiger W (1990) Detection of some conserved domains in phytochrome-like proteins from algae. J Plant Physiol 136:484–487
Martin W, Gierl A, Saedler H (1989) Molecular evidence for preCretaceous angiosperm origins. Nature 339:46–48
Martin W, Sommerville CC, Loiseaux-de Goër (1992) Molecular phylogenies of plastid origins and algal evolution. J Mol Evol 35:385–403
Martin W, Lydiate D, Brinkmann H, Forkman G, Saedler H, Cerff R (1993) Molecular phylogenies in angiosperm evolution. Mol Biol Evol 10:140–162
Maucher HP, Scheuerlein R, Schraudolf H (1992) Detection and partial sequence of phytochrome genes in the fernAnemia phyllitidis (L.) Sw (Schizaeaceae) andDryopteris filix-mas L. (Polypodiaceae) by using polymerase-chain reaction technology. Photochem Photobiol 56:759–763
Neuhaus G, Bowler C, Kern R, Chua N-H (1993) Calcium/calmodulindependent and -independent phytochrome signal transduction pathways. Cell 73:937–952
Okamoto H, Hirano Y, Abe H, Tomizawa KI, Furuya M, Wada M (1993) The deduced amino acid sequence of phytochrome fromAdiantum includes consensus motifs present in phytochrome B from seed plants. Plant Cell Physiol 34:1329–1334
Ota T, Nei M (1994) Estimation of the number of amino acid substitutions per site when the substitution rate varies among sites. J Mol Evol 38:642–643
Raubeson LA, Jansen RK (1992) Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 255: 1697–1699
Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions inArabidopsis development. Plant Physiol 104:1139–1149
Rüdiger W, Thümmler F (1991) Phytochrome, the visual pigment of plants. Angew Chem Int Ed Engl 30:1216–1228
Saccone C, Lanave C, Pesole G (1993) Time and biosequences. J Mol Evol 37:154–159
Sarich VM, Wilson AC (1973) Generation time and genomic evolution in primates. Science 179:1144–1147
Schneider-Poetsch HAW (1992) Signal transduction by phytochrome: phytochromes have a module related to the transmitter modules of bacterial sensor proteins. Photochem Photobiol 56:839–846
Schneider-Poetsch HAW, Braun B, Rüdiger W (1989) Phytochromeall regions marked by a set of monoclonal antibodies reflect conformational changes. Planta 177:511–514
Schneider-Poetsch HAW, John G, Braun B (1990) The distribution of a phytochrome-like protein in the fernPsilotum nudum. An immunoblotting analysis of an early ancestor of all vascular plants. Bot Acta 103:235–239
Schneider-Poetsch HAW, Marx S, Kolukisaoglu HO, Hanelt S, Braun B (1994) Phytochrome evolution: Phytochrome genes in ferns and mosses. Physiol Plant 91:241–250
Sharrock RA, Quail HP (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution and differential expression of a plant regulatory photoreceptor family. Genes Dev 3:1745–1757
Shinomura T, Nagatani A, Chory J, Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104:363–371
Sitte P, Ziegler H, Ehrendorfer F, Bresinsky A (1991) Strasburger—Lehrbuch der Botanik. G Fischer, Stuttgart
Stein DB, Conant DS, Abeam ME, Jordan ET, Kirch SA, Hasebe M, lwatsuki K, Tan MK, Thomson JA (1992) Structural rearrangements of the chloroplast genome provide an important phylogenetic link in ferns. Proc Natl Acad Sci USA 89:1856–1860
Stewart WN, Rothwell GW (1993) Paleobotany and the evolution of plants, 2nd ed. Cambridge University Press, Cambridge
Thümmler F, Dufner M, Dittrich P (1992) Molecular cloning of a novel phytochrome gene of the mossCeratodon purpureus which encodes a putative light-regulated protein kinase. Plant Mol Biol 20: 1003–1017
Winands A, Wagner G, Marx S, Schneider-Poetsch HAW (1992) Partial nucleotide sequence of the Zygnematophycean green algaMougeotia. Photochem Photobiol 56:765–770
Wolfe KH, Gouy M, Yang YW, Sharp P, Li W-H (1989) Date of the monocotdicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86:6201–6205
Zharkikh A, Li W-H (1992) Statistical properties of bootstrap estimation of phylogenetic variability from nucleotide sequences. II. Four taxa without a molecular clock. J Mol Evol 35:356–366
Author information
Authors and Affiliations
Additional information
Correspondence to: H.A.W. Schneider-Poetsch
Rights and permissions
About this article
Cite this article
Kolukisaoglu, H.Ü., Marx, S., Wiegmann, C. et al. Divergence of the phytochrome gene family predates angiosperm evolution and suggests thatSelaginella andEquisetum arose prior toPsilotum . J Mol Evol 41, 329–337 (1995). https://doi.org/10.1007/BF01215179
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01215179