Abstract
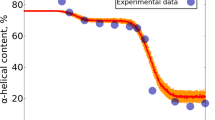
To test the hypothesis that the folding pathways of evolutionarily related proteins with similar three-dimensional structures but widely different sequences should be similar, the folding pathway of apoleghemoglobin has been characterized using stopped-flow circular dichroism, heteronuclear NMR pulse labeling techniques and mass spectrometry. The pathway of folding was found to differ significantly from that of a protein of the same family, apomyoglobin, although both proteins appear to fold through helical burst phase intermediates. For leghemoglobin, the burst phase intermediate exhibits stable helical structure in the G and H helices, together with a small region in the center of the E helix. The A and B helices are not stabilized until later stages of the folding process. The structure of the burst phase folding intermediate thus differs from that of apomyoglobin, in which stable helical structure is formed in the A, B, G and H helix regions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lesk, A.M. & Chothia, C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J. Mol. Biol. 136, 225–270 (1980).
Bashford, D., Chothia, C. & Lesk, A.M. Determinants of a protein fold. Unique features of the globin amino acid sequence. J. Mol. Biol. 196, 199–216 (1987).
Hollecker, M. & Creighton, T.E. Evolutionary conservation and variation of protein folding pathways. J. Mol. Biol. 168, 409–437 (1983).
Krebs, H., Schmid, F.X. & Jaenicke, R. Folding of homologous proteins. J. Mol. Biol. 169, 619–635 (1983).
Stackhouse, T.M., Onuffer, J.J., Matthews, C.R., Ahmed, S.A. & Miles, E.W. Folding of homologous proteins: conservation of the folding mechanism of the a subunit of tryptophan synthase from Escherichia coli, Salmonella typhimurium, and five interspecies hybrids. Biochemistry 27, 824–832 (1988).
Jaenicke, R. Folding and association of proteins. Prog. Biophys. Mol. Biol. 49, 117–237 (1987).
Plaxco, K.W., Spitzfaden, C., Campbell, I.D. & Dobson, C.M. A comparison of the folding kinetics and thermodynamics of two homologous fibronectin type III modules. J. Mol. Biol. 270, 763–770 (1997).
Chiti, F., et al. Mutational analysis of acylphosphatase suggests the importance of topology and contact order in protein folding. Nature Struct. Biol. 6, 1005–1009 (1999).
Riddle, D.S., Grantcharova, V.P., Santiago, J.V., Alm, E., Ruczinski, I. & Baker, D. Experiment and theory highlight role of native state topology in SH3 folding. Nature Struct. Biol. 6, 1016–1024 (1999).
Martinez, J.C. & Serrano, L. The folding transition state between SH3 domains is conformationally restricted and evolutionarily conserved. Nature Struct. Biol. 6, 1010–1016 (1999).
Ptitsyn, O.B. & Ting, K.L. Non-functional conserved residues in globins and their possible role as a folding nucleus. J. Mol. Biol. 291, 671–682 (1999).
Appleby, C.A. Leghemoglobin and Rhizobium respiration. Annu. Rev. Plant Physiol. 35, 443–478 (1984).
Appleby, C.A. The origin and functions of hemoglobin in plants. Sci. Progr. 76, 365–398 (1992).
Landsmann, J., Dennis, E.S., Higgins, T.J.V., Appleby, C.A., Kortt, A.A. & Peacock, W.J. Common evolutionary origin of legume and non-legume plant haemoglobins. Nature 324, 166–168 (1986).
Callaway, D.J.E. Solvent-induced organization: a physical model of folding myoglobin. Proteins 20, 124–138 (1994).
Chelvanayagam, G., Reich, Z., Bringas, R. & Argos, P. Prediction of protein folding pathways. J. Mol. Biol. 227, 901–916 (1992).
Hargrove, M.S., et al. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J. Mol. Biol. 266, 1032–1042 (1997).
Ellis, P.J., Appleby, C.A., Guss, J.M., Hunter, W.N., Ollis, D.L. & Freeman, H.C. The crystal structure of ferric soybean leghemoglobin a nicotinate at 2.3 Å resolution. Acta Crystallogr. D 53, 302–310 (1997).
Kuriyan, J., Wilz, S., Karplus, M. & Petsko, G.A. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5 Å resolution. J. Mol. Biol. 192, 133–154 (1986).
Morikis, D. & Wright, P.E. Hydrogen exchange in the carbon monoxide complex of soybean leghemoglobin. Eur. J. Biochem. 237, 212–220 (1996).
Tsui, V., Garcia, C., Cavagnero, S., Siuzdak, G., Dyson, H.J. & Wright, P.E. Quench-flow experiments combined with mass spectrometry show apomyoglobin folds through an obligatory intermediate. Protein Sci. 8, 45–49 (1999).
Jennings, P.A. & Wright, P.E. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science 262, 892–896 (1993).
Hughson, F.M., Wright, P.E. & Baldwin, R.L. Structural characterization of a partly folded apomyoglobin intermediate. Science 249, 1544–1548 (1990).
Eliezer, D., Yao, J., Dyson, H.J. & Wright, P.E. Structural and dynamic characterization of partially folded states of myoglobin and implications for protein folding. Nature Struct. Biol. 5, 148–155 (1998).
Jamin, M. & Baldwin, R.L. Refolding and unfolding kinetics of the equilibrium folding intermediate of apomyoglobin. Nature Struct. Biol. 3, 613–618 (1996).
Luo, Y., Kay, M.S. & Baldwin, R.L. Cooperativity of folding of the apomyoglobin pH 4 intermediate studied by glycine and proline mutations. Nature Struct. Biol. 4, 925–930 (1997).
Cavagnero, S., Dyson, H.J. & Wright, P.E. Effect of H helix destabilizing mutations on the kinetic and equilibrium folding of apomyoglobin. J. Mol. Biol. 285, 269–282 (1999).
Garcia, C., Nishimura, C., Cavagnero, S., Dyson, H.J. & Wright, P.E. Changes in the apomyoglobin folding pathway caused by mutations of the distal histidine residue. Biochemistry in the press (2000).
Rose, G.D., Geselowitz, A.R., Lesser, G.J., Lee, R.H. & Zehfus, M.H. Hydrophobicity of amino acid residues in globular proteins. Science 229, 834–838 (1985).
Muñoz, V. & Serrano, L. Development of the multiple sequence approximation within the AGADIR model of α-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers 41, 495–509 (1997).
Waltho, J.P., Feher, V.A., Merutka, G., Dyson, H.J. & Wright, P.E. Peptide models of protein folding initiation sites. 1. Secondary structure formation by peptides corresponding to the G- and H-helices of myoglobin. Biochemistry 32, 6337–6347 (1993).
Shin, H.-C., Merutka, G., Waltho, J.P., Tennant, L.L., Dyson, H.J. & Wright, P.E. Peptide models of protein folding initiation sites. 3. The G-H helical hairpin of myoglobin. Biochemistry 32, 6356–6364 (1993).
Dalessio, P.M. & Ropson, I.J. β-sheet proteins with nearly identical structures have different folding intermediates. Biochemistry 39, 860–871 (2000).
Prytulla, S., Dyson, H.J. & Wright, P.E. Gene synthesis, high-level expression and assignment of backbone 15N and 13C resonances of soybean leghemoglobin. FEBS Lett. 399, 283–289 (1996).
Ellfolk, N. & Sievers, G. Circular dichroism of soybean leghemoglobin. Biochim. Biophys. Acta 405, 213–227 (1975).
Delaglio, F., Grzesiek, S., Vuister, G.W., Guang, Z., Pfeifer, J. & Bax, A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Morikis, D., Lepre, C.A. & Wright, P.E. 1H resonance assignments and secondary structure of the carbon monoxide complex of soybean leghemoglobin determined by homonuclear two-dimensional and three-dimensional NMR spectroscopy. Eur. J. Biochem. 219, 611–626 (1994).
Miranker, A., Robinson, C.V., Radford, S.E. & Dobson, C.M. Investigation of protein folding by mass spectrometry. FASEB J. 10, 93–101 (1996).
Cavagnero, S., Thériault, Y., Narula, S.S., Dyson, H.J. & Wright, P.E. Amide proton hydrogen exchange rates for sperm whale myoglobin obtained from 15N-1H NMR spectra. Protein Sci. 9, 186–193 (2000).
Acknowledgements
We thank J. Chung for assistance with the NMR experiments, D. Eliezer, J. Yao, P. Jennings, C. Garcia, V. Tsui and S. Cavagnero for helpful discussions, and L. Tennant for expert technical assistance. This work was supported by grants from the National Institutes of Health. S.P. was supported by a fellowship from the Deutsches Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Nishimura, C., Prytulla, S., Jane Dyson, H. et al. Conservation of folding pathways in evolutionarily distant globin sequences. Nat Struct Mol Biol 7, 679–686 (2000). https://doi.org/10.1038/77985
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/77985
This article is cited by
-
Exploring the Sequence-based Prediction of Folding Initiation Sites in Proteins
Scientific Reports (2017)
-
Sequence analysis on the information of folding initiation segments in ferredoxin-like fold proteins
BMC Structural Biology (2014)
-
Predicting protein folding pathways at the mesoscopic level based on native interactions between secondary structure elements
BMC Bioinformatics (2008)
-
Mechanisms of protein folding
European Biophysics Journal (2008)
-
Apomyoglobin stability as dependent on urea concentration and temperature at two pH values
Molecular Biology (2005)